Introduction to Pan Genome
A pan-genome (pangenome or supragenome) is the whole set of genes from all strains within a clade in molecular biology and genetics. It’s the sum of a clade’s genomes in a broader perspective. The pan-genome is divided into three sections: a “core pangenome” that contains genes found in all individuals, a “shell pangenome” that contains genes found in two or more strains, and a “cloud pangenome” that only has genes seen in one strain. Some authors refer to the cloud genome as an “accessory genome,” which contains “dispensable” genes found in only a subset of strains and strain-specific genes. At least in plant genomes, the term “dispensable” has been questioned, as accessory genes “play an important role in genome evolution and the complex interplay between the genome and the environment.”
The advent of next-generation sequencing allowed for a more in-depth examination of the diversity of several bacterial strains, thereby expanding our understanding of the concept of species. Following that, comparative studies between members of the same species or genus have become the most common method for identifying unique genes and genes shared between two or more species, as well as characterizing all genes shared among all individuals (homologous genes) evaluated. This method is known as pan-genome, and it currently employs a variety of bioinformatics tools as well as a variety of computational approaches to identify genomic data and the set of genes that represent a given taxon. Pan-genome studies and their various techniques have even been utilized in eukaryotes, particularly humans, in order to better comprehend the genetic variability that exists between individuals.
Sequencing Methods Used in Pangenomic Studies
Single Molecule Real-Time (SMRT) Sequencing and Nanopore Sequencing are currently commercializing single-molecule, long-read technologies. SMRT Sequencing enables (1) cultivation pan-genomes to classify the complete genetic diversity within a species, (2) innovation of population-specific reference genomes to propel precision medicine and (3) configuration of near-complete microbial genomes and their plasmids in a single experiment using whole genome sequencing for de novo assembly. While methodologies based on short-read sequencing approaches are also being established to offer long-range single-molecule level information—for example, the 10x Genomics linked read methodology.
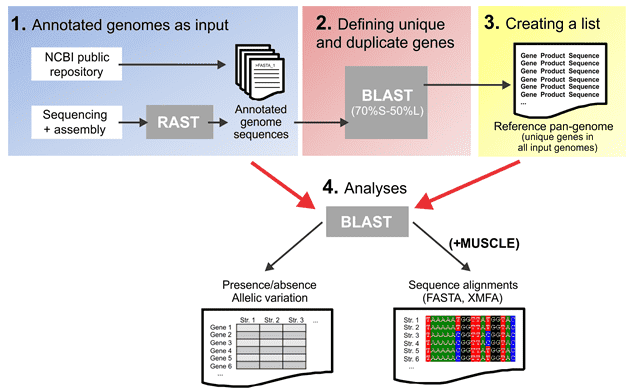
Figure 1. A Reference Pan-Genome Approach to Comparative Bacterial Genomics. (Méric, 2014)
Applications of Pangenome
For pan-genomic analysis, a variety of bioinformatics tools have been used, which could help define species. It may be feasible to redefine species and sort them based on their genomic content by analyzing all genomic data. Since the last decade, pan-genomic analyses have paved the way for researchers to create universal vaccines that could protect against all strains in a species or even against several related species.
The field of pathogenic pan-genomics has been revolutionized by recent advances in genome sequencing technology, which has also influenced disease management in aqua farms. The phylogenomic diversity and possible evolutionary trends of aquatic bacterial pathogen strains will be deduced through routine pan-genome analysis of genomic-derived aquatic pathogens, as well as the mechanisms of pathogenesis, even the estimated patterns of pathogen transmission across epidemiological scales.
References:
- Sherman RM, Salzberg SL. Pan-genomics in the human genome era. Nature Reviews Genetics. 2020 Apr;21(4).
- Tiwary BK. Evolutionary pan-genomics and applications. InPan-genomics: Applications, Challenges, and Future Prospects 2020 Jan 1. Academic Press.
- Méric G, Yahara K, Mageiros L, et al. A reference pan-genome approach to comparative bacterial genomics: identification of novel epidemiological markers in pathogenic Campylobacter. PloS one. 2014 Mar 27;9(3).

